 I’ve been following Razib Khan’s scholarly and analytical exploration of his family’s genetic history – using data from 23andMe – over at Gene Expression with increasing fascination. When last week he noted that his findings appeared to be (finally) converging on a consensus, I asked if he’d be willing to summarise his journey for Genomes Unzipped readers. Here it is. –DM.
I’ve been following Razib Khan’s scholarly and analytical exploration of his family’s genetic history – using data from 23andMe – over at Gene Expression with increasing fascination. When last week he noted that his findings appeared to be (finally) converging on a consensus, I asked if he’d be willing to summarise his journey for Genomes Unzipped readers. Here it is. –DM.
I’ve always been interested in genetics, anthropology, and history. Many may perceive me to be a collector of obscure facts, but summing up infinitesimals does produce something substantial in aggregation. One of the most influential books in my life has been History and Geography of Genes. So with that, the shift from classical markers to uniparental lineages, and now to the dense SNP-chips, has been a boon for my own intellectual interests which reside in part at the intersection of history and population genetics.
However, I’ve never been deeply curious as to the history of my own personal genome. I’m not adopted. All four of my grandparents were ethnic Bengalis, albeit from relatively diverse communal backgrounds. I look typically South Asian. Genealogy has never been a family fascination, and I’m going to be honest and admit that until five years ago I didn’t even know the names of my grandparents (in the Bengali language there are distinctive terms for maternal and paternal grandparents, so this wasn’t needed). Both sides of my family are from the Comilla district of Bengal, and that’s all I really cared about (and I didn’t care that much, I don’t put much stock in “heritage” as determinative).
As for other yields of personal genomics, I was skeptical. My parents have many siblings, and many, many, cousins. I had a general sense of my risks for diseases through an inspection of the pedigree of my family and their medical histories. Additionally, many of the risk alleles have been identified in European study populations, and I wasn’t totally sure about the between-population portability of these inferences. And I won’t even address the fact that effect size of many of the markers isn’t something to shout home about.
 But last spring Daniel alerted me to the 23andMe “DNA Day” sale. It was affordable, and at that point enough of the readers of my weblog had been typed that I kept getting questions as to my own background (e.g., my family has the title Khan, so there was a question as to whether I carried the “Genghis Khan haplotype”). So I bit. At the time I recall emailing Dan and being excited that I’d be told I likely had brown eyes and was 75% “European” and 25% “Asian.” When my results came back, I was in for a mild surprise. The proportion to the left are calculated by 23andMe’s “ancestry painting” algorithm. As you can see, I’m more than 25% “Asian.” My initial reaction was that this seemed a touch high, but no worries, I would ask around and see which other South Asians had such a high value. After dozens of instances of “gene sharing,” the answer came back: none.
But last spring Daniel alerted me to the 23andMe “DNA Day” sale. It was affordable, and at that point enough of the readers of my weblog had been typed that I kept getting questions as to my own background (e.g., my family has the title Khan, so there was a question as to whether I carried the “Genghis Khan haplotype”). So I bit. At the time I recall emailing Dan and being excited that I’d be told I likely had brown eyes and was 75% “European” and 25% “Asian.” When my results came back, I was in for a mild surprise. The proportion to the left are calculated by 23andMe’s “ancestry painting” algorithm. As you can see, I’m more than 25% “Asian.” My initial reaction was that this seemed a touch high, but no worries, I would ask around and see which other South Asians had such a high value. After dozens of instances of “gene sharing,” the answer came back: none.
Continue reading ‘Guest post by Razib Khan: My personal genome’

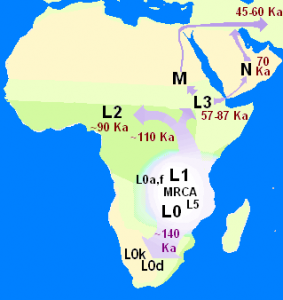
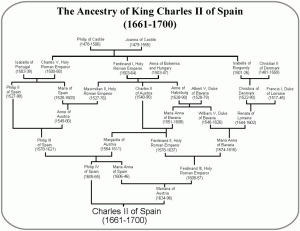
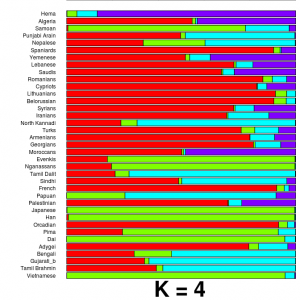

 But last spring Daniel alerted me to the 23andMe “DNA Day” sale. It was affordable, and at that point enough of the readers of my weblog had been typed that I kept getting questions as to my own background (e.g., my family has the title Khan, so there was a question as to whether I carried the “Genghis Khan haplotype”). So I bit. At the time I recall emailing Dan and being excited that I’d be told I likely had brown eyes and was 75% “European” and 25% “Asian.” When my results came back, I was in for a mild surprise. The proportion to the left are calculated by 23andMe’s “ancestry painting” algorithm. As you can see, I’m more than 25% “Asian.” My initial reaction was that this seemed a touch high, but no worries, I would ask around and see which other South Asians had such a high value. After dozens of instances of “gene sharing,” the answer came back: none.
But last spring Daniel alerted me to the 23andMe “DNA Day” sale. It was affordable, and at that point enough of the readers of my weblog had been typed that I kept getting questions as to my own background (e.g., my family has the title Khan, so there was a question as to whether I carried the “Genghis Khan haplotype”). So I bit. At the time I recall emailing Dan and being excited that I’d be told I likely had brown eyes and was 75% “European” and 25% “Asian.” When my results came back, I was in for a mild surprise. The proportion to the left are calculated by 23andMe’s “ancestry painting” algorithm. As you can see, I’m more than 25% “Asian.” My initial reaction was that this seemed a touch high, but no worries, I would ask around and see which other South Asians had such a high value. After dozens of instances of “gene sharing,” the answer came back: none. RSS
RSS Twitter
Twitter
Recent Comments