 Those of us involved in genomics research spend a lot of time thinking about how scientific and technological developments might influence personal genomics. For instance, does the falling cost of sequencing mean that medically useful personal genomics will likely be based on sequence rather than genotype data? (Yes.)
Those of us involved in genomics research spend a lot of time thinking about how scientific and technological developments might influence personal genomics. For instance, does the falling cost of sequencing mean that medically useful personal genomics will likely be based on sequence rather than genotype data? (Yes.)
At the Sanger Institute we’ve recently launched (along with our friends at EBI) a project to look more deeply at a question which is less often on the lips of genomics boffins: “How does genomics affect as us people, both individually and in communities?” Because of the obvious resonance with Genomes Unzipped it should come as no surprise that many of us (including myself, Daniel and Luke) have been intimately involved in this initiative.
The actual line-up of events has been diverse, and a lot of fun. We’ve had two excellent debates, including one between Ewan Birney and Paul Flicek (pictured) on the value, or lack thereof, of celebrity genomes (covered in more detail here). A poet, Fiona Sampson, spent some time on campus and we’ve commissioned a book of poetry from her. This one raised some eyebrows, but I have to say that talking to her has given me some brand new ways of thinking about my own work. We’re also working on a more interactive project in the hope of making personal genomics a bit more personal. Stay tuned.
 Last week, a post went up on the Bioscience Resource Project blog entited The Great DNA Data Deficit. This is another in a long string of “Death of GWAS” posts that have appeared around the last year. The authors claim that because GWAS has failed to identify many “major disease genes”, i.e. high frequency variants with large effect on disease, it was therefore not worthwhile; this is all old stuff, that I have discussed elsewhere (see also my “Standard GWAS Disclaimer” below). In this case, the authors argue that the genetic contribution to complex disease has been massively overestimated, and in fact genetics does not play as large a part in disease as we believe.
Last week, a post went up on the Bioscience Resource Project blog entited The Great DNA Data Deficit. This is another in a long string of “Death of GWAS” posts that have appeared around the last year. The authors claim that because GWAS has failed to identify many “major disease genes”, i.e. high frequency variants with large effect on disease, it was therefore not worthwhile; this is all old stuff, that I have discussed elsewhere (see also my “Standard GWAS Disclaimer” below). In this case, the authors argue that the genetic contribution to complex disease has been massively overestimated, and in fact genetics does not play as large a part in disease as we believe.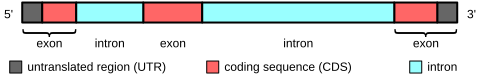
 RSS
RSS Twitter
Twitter
Recent Comments