 As I mentioned a few weeks ago, we recently published a large study into the genetics of inflammatory bowel disease (IBD), which included a number of analyses digging into the biology and evolutionary history of IBD genetic risk. Gratifyingly, our paper has stimulated a lot of discussion among other scientists, which has generated several ideas about future directions for this work. One question that was raised by several population-genetics experts at ASHG was about our natural selection analysis, and in particular our claim to discover an enrichment of balancing selection in IBD loci. In the paper, we found clear signals of natural selection on IBD loci, a subset of which we interpreted as balancing selection. In this post I will set out how I came to this conclusion, but then outline another explanation that could explain the results: recent local positive selection in Europeans.
As I mentioned a few weeks ago, we recently published a large study into the genetics of inflammatory bowel disease (IBD), which included a number of analyses digging into the biology and evolutionary history of IBD genetic risk. Gratifyingly, our paper has stimulated a lot of discussion among other scientists, which has generated several ideas about future directions for this work. One question that was raised by several population-genetics experts at ASHG was about our natural selection analysis, and in particular our claim to discover an enrichment of balancing selection in IBD loci. In the paper, we found clear signals of natural selection on IBD loci, a subset of which we interpreted as balancing selection. In this post I will set out how I came to this conclusion, but then outline another explanation that could explain the results: recent local positive selection in Europeans.
(I am going to use “I” rather than “we” from here on despite the large number of co-authors on the paper, because the selection analysis (with its flaws) was entirely mine.)
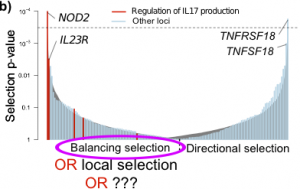
First let’s review what was actually in the paper: to investigate selection on IBD loci I used the data generated using TreeMix and the HGDP data by fellow GNZ author Joe Pickrell. I found tag SNPs for 162 of the IBD risk variants in the HGDP data, and calculated p-values for each by comparing its selection score to scores from a frequency-matched set of SNPs. I looked for SNPs that were extreme in either direction, i.e. with either higher or lower selection scores. By analogy to Tajima’s D, I characterised these two extremes as “directional” and “balancing” selection, based on the hypothesis that direction selection will lead to deviations from the population tree, but balancing selecting will lead to variants that are under-dispersed relative to the tree. The “balancing” interpretation was an “off-label” use of the TreeMix method, which Joe had previously used to study only directional selection.
I found significant signal at both ends: IBD risk loci were far more likely to have both higher (“directional”) and lower (“balancing”) scores values than the sets of frequency-matched SNPs. The strongest “balancing” selection signal was also the strongest association signal (NOD2), which had previously been identified as showing evidence for balancing selection. Furthermore, the variants that showed “balancing” selection were more likely than other IBD risk loci to be near genes in certain functional categories, including IL-17 regulation and response to bacteria. As these functions are good categories for balancing selection I felt confident in concluding that the “balancing” selection signal I saw was indeed a signal of balancing selection.
In retrospect, however, I may have come to this conclusion too hastily, and too naively. Since the paper was published, several population genetics colleagues (including Joe, whose data I was using), have pointed out that while some models of balancing selection may lead to variants hugging closely to the tree, many others will not. Furthermore, they have expressed some skepticism that this method can detect realistic signals of balancing selection due to the (relatively) short human time frames involved.
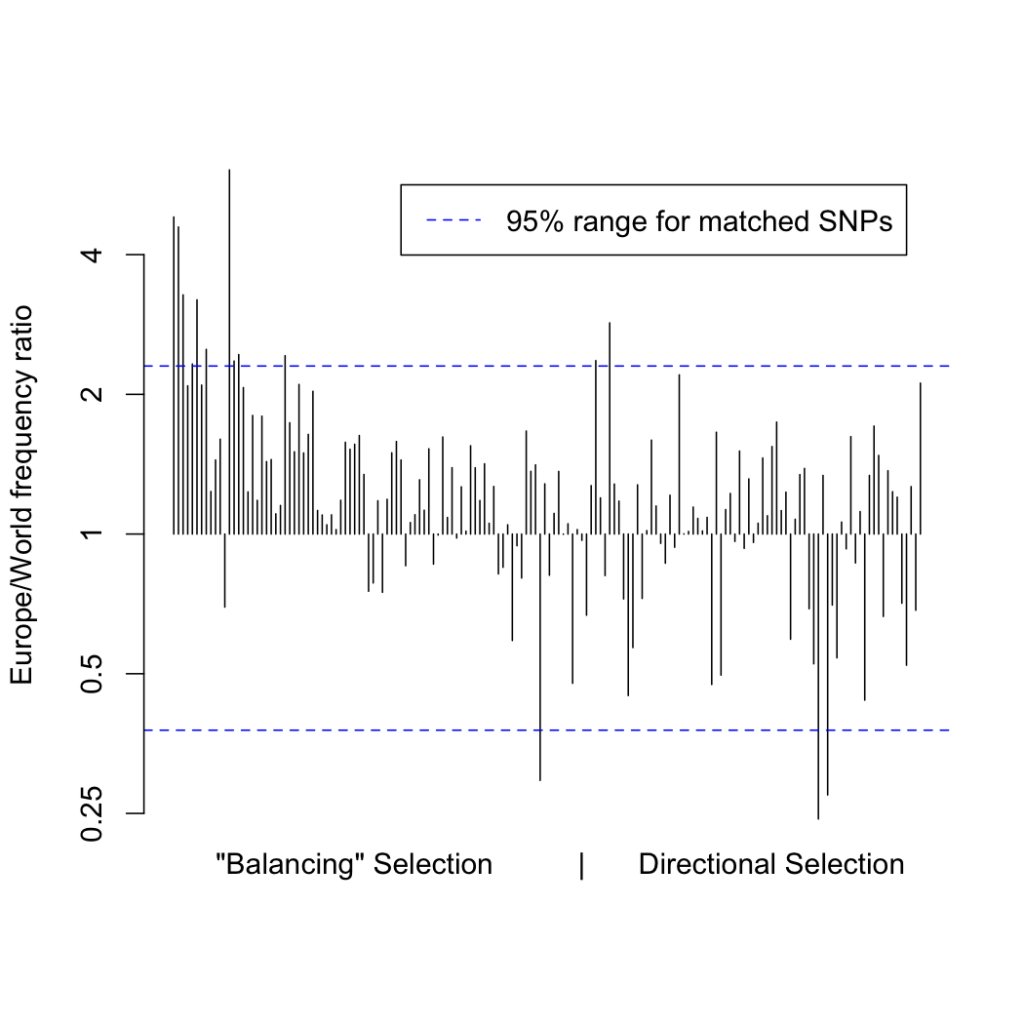
In response to these criticisms, I went back to the data to see if any other effect could explain the signal I reported. Digging deeper into the frequency distribution of the variants under selection, I noticed a particular pattern: the variants that I had described as under “balancing selection” were at systematically higher frequency in Europe compared to the rest of the world than other variants matched for European allele frequency:
I normalized for this in the initial analysis for the exact reason that the selection score is known to be confounded by frequency. However, I only normalized by frequency in Europeans, as this is where the biases due to our GWAS discovery method would manifest. Instead, if we consider normalizing world-wide, we can see that while the positive selection p-values are robust to the normalizing method, the “balancing” selection score is not:
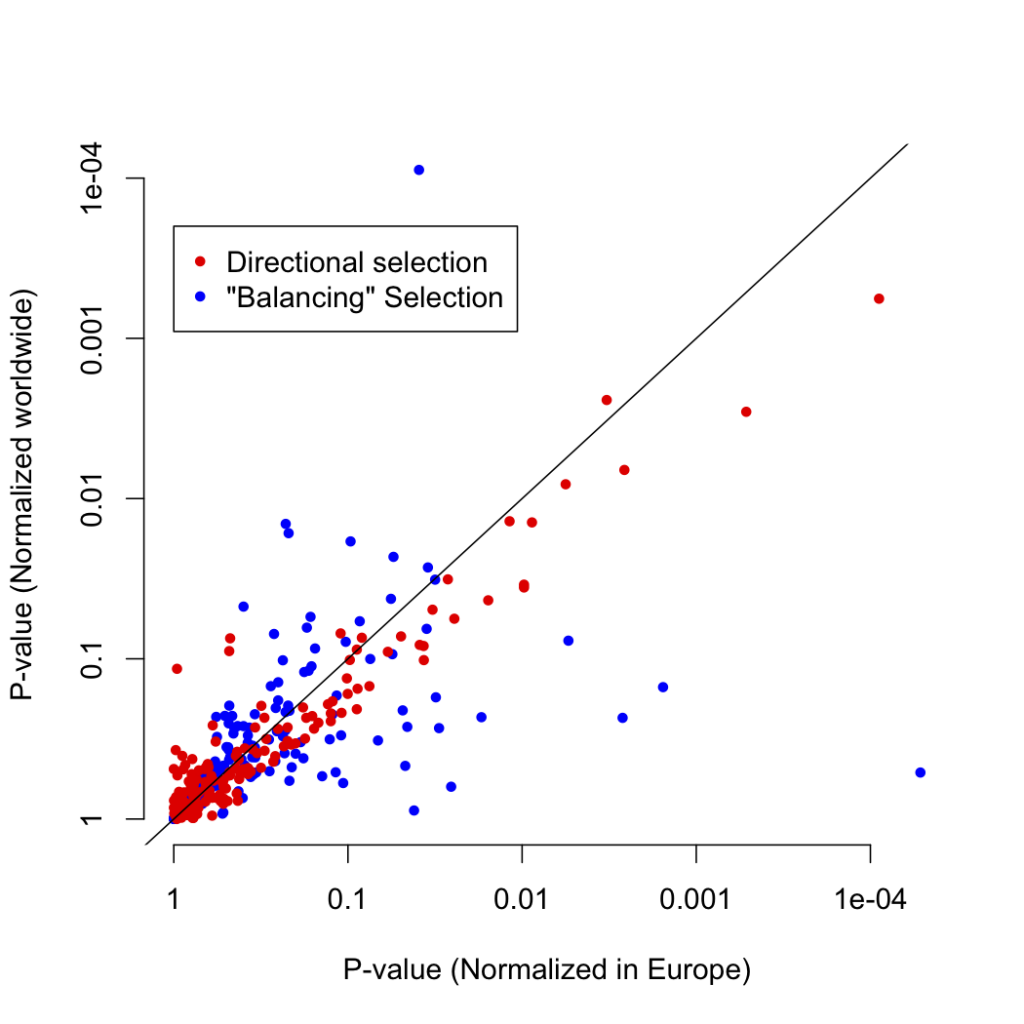
This leads us to the conclusion that rather than being caused by underdispersed variants, the “balancing” selection signal is driven by IBD risk variants with abnormally low frequencies outside of Europe. In the absence of selection of some kind, we would expect our IBD risk loci to have the same frequency spectrum outside Europe as SNPs matched on European frequency. However, instead of balancing selection, this signal could also be explained by recent positive selection in Europeans that was stronger than in other populations.
The hypothesis of recent local selection on IBD regions explains the observed data as well as, or better than, balancing selection. That these expanded IBD loci would preferentially show evidence of pathways involved in interacting with bacteria also makes sense, given the hypothesized importance of settled farming life-styles during the Neolithic expansion. However, I have learned my lesson about speculating too soon about selection! So instead I will just note that we have shown that IBD risk variants show evidence of two different forms of selection, and leave untangling exactly what these are to further research.
For me, this experience has reinforced the value of communicating with relevant experts after or, perhaps preferably, before publication. If I had discussed this analysis with Joe prior to publishing it, I doubtless would have modified the claims I made in the paper. Errors or omissions are mostly likely to be made when people make claims outside their direct area of expertise. Finding the time to discuss ideas with those in the know, even when time is tight and you have many analyses on the go, can make science run a lot smoother.
Thank you to all the population geneticists who commented on the paper, including Joe Pickrell, Graham Coop and Gil McVean.








 RSS
RSS Twitter
Twitter
bravo!’tis a far better thing to admit an aspect of error if it brings us closer to the mark of truth. and yet this is a rare act indeed.
Hi Luke,
Nice to see your thoughts on this. A few further thoughts/points of clarification.
The reason that alleles that are under-dispersed are hard to detect is that for low differentiation there are many loci that have only small changes in frequency between populations. If we look at the distribution of Fst across loci, and Fst is not very large, this distribution is L shaped and there are many loci with near zero Fst. This means that a test that looks for under-dispersed loci is under-powered, when populations are only recently diverged, and tests of extreme differentiation are much more powerful (e.g. see here). Over longer time-scales we could hope to see loci that maintained polymorphism, and indeed looking for trans-specific polymorphism is one test of balancing selection (and conversely tests of extreme differentiation are very underpowered as many loci are fixed differences).
That said, the idea that balancing selection leads to strongly under-dispersed allele frequencies among populations has long seemed strange to me. Strong heterozygote advantage (or other simple negative frequency dependent models) would keep the allele frequencies relatively constant, if the relative fitnesses of the genotypes stayed constant. However, it seems likely that these fitnesses will vary across environments. Also under more general models of balancing selection, temporal or spatially selection, we are invoking variation across conditions to maintain polymorphism and so we shouldn’t expect frequencies to stay constant.
We’ve developed a test for loci that strongly differ from a covariance matrix of allele frequencies (see here), that is along the lines of the one suggested by Joe but that has some nice features that make it a close analog of Fst. Once again I think this is better at detecting overly differentiated loci than balanced loci [but our tests of environmental correlations may well be a good way of finding spatial balanced polymorphisms].
Anyway great to see your thoughts on this, and hope this helps.
Graham