Readers who don’t have a Science subscription can access a pre-formatted version of the manuscript here. In this post I wanted to give a brief overview of the study and then highlight what I see as some of the interesting messages that emerged from it.
First, some background
This is a project some three years in the making – the idea behind it was first conceived by my Sanger colleague Bryndis Yngvadottir and I back in 2009, and it subsequently expanded into a very productive collaboration with several groups, most notably Mark Gerstein’s group at Yale University, and the HAVANA gene annotation team at the Sanger Institute.
The idea is very simple. We’re interested in loss-of-function (LoF) variants – genetic changes that are predicted to be seriously disruptive to the function of protein-coding genes. These come in many forms, ranging from a single base change that creates a premature stop codon in the middle of a gene, all the way up to massive deletions that remove one or more genes completely. These types of DNA changes have long been of interest to geneticists, because they’re known to play a major role in really serious diseases like cystic fibrosis and muscular dystrophy.
But there’s also another reason that they’re interesting, which is more surprising: every complete human genome sequenced to date, including celebrities like James Watson and Craig Venter, has appeared to carry hundreds of these LoF variants. If those variants were all real, that would indicate a surprising degree of redundancy in the human genome. But the problem is we don’t actually know how many of these variants are real – no-one has ever taken a really careful look at them on a genome-wide scale.
So that’s what we did. Working with the pilot data from the 1000 Genomes Project – whole-genome sequences from 185 individuals with ancestry from Europe, East Asia and West Africa – we made a list of nearly 3,000 predicted LoF variants found in these genomes, and put them through a punishing barrage of experimental validation and computational filtering. In addition, we were fortunate enough to have the HAVANA team on our side, and their annotators (coordinated by Adam Frankish) spent hundreds of man-hours manually checking the annotations underlying about 700 genes containing these predicted variants. At the end of that frankly pretty exhausting process we were left with a grand total of 1,285 variants that we think probably result in genuine loss of function.
Dysfunctional genomes
So we were left with a core set of “real” LoF variants – now we could start doing some interesting biology.
Firstly, after applying every filter we could think of, we ended up with a fairly stable estimate of the true number of LoF variants per individual – we think each of us (at least those of us of non-African ancestry) carries about 100 of these variants, with 20 of those being present in a homozygous state (i.e. both copies of the affected gene are inactivated). These numbers are perhaps a little higher in Africans, consistent with their overall higher levels of genetic variation.
A relatively small number (253) of these LoF variants were quite common, with at least one individual in the study having both copies knocked out. Some of these are pretty interesting: we found known LoF variants associated with things like blood type, muscle performance and drug metabolism, for instance, along with a vast array of inactivated olfactory receptors (genes involved in smelling particular substances). However, we found little evidence for a role of these common LoF variants in risk for common, complex diseases like type 2 diabetes, despite a frankly heroic effort by GNZ comrade Luke Jostins to track these down (for those in the know: imputing the entire 1000 Genomes data-set into all 17,000 people in the WTCCC1 study – yeah, that isn’t easy). That fits with what we saw when we looked at the affected genes: those hit by common LoF variants tended to be less evolutionarily conserved, had fewer known protein-protein interactions, and were also more likely to have other similar genes in the genome. That suggests that in general these genes are more redundant and less functionally critical: no surprise, given they’re knocked out in a non-trivial fraction of the population.
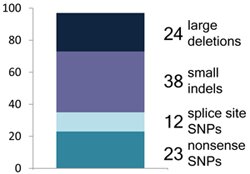
But overall, we found that genuine LoF variants tend to be extremely rare: the majority of our high-confidence LoF variants were found in less than 2% of the population, with many probably being far rarer than that. This suggests that LoF variants are often mildly or severely deleterious, and have thus been stopped from increasing in frequency by natural selection. That in turn suggests that those rare LoF variants are where most of the action is in terms of effects on disease risk. Sure enough, we found 24 known severe disease-causing mutations in our LoF set, involved in horrible diseases like osteogenesis imperfecta and harlequin ichthyosis, as well as 21 LoF variants in known disease-causing genes – these were all found in only one copy in the affected individuals.
There’s more, including an analysis of the effects of these variants on RNA expression (courtesy in part of GNZ’s own Joe Pickrell), and an algorithm we think might be useful for predicting whether a novel mutation is likely to actually cause disease – but I’m going to skim over those, and instead do my best to hammer home a point about error rates that I think is absolutely critical to appreciate for those currently setting up their own sequencing projects.
The more interesting something is, the less likely it is to be real
We are rapidly entering a world in which sequencing genomes is becoming commonplace. It is not unusual now for a PhD student to contemplate a project in which the complete sequences of the protein-coding genes (the exomes) of dozens of disease patients will be examined. Over the next 12 months tens of thousands of disease patients and healthy controls will have their exomes or complete genomes sequenced.
As the data roll off those projects, researchers will naturally find their eyes drawn to the genetic variants with the largest predicted effects on function – in other words, the ones most likely to be involved in disease. And while the smart researchers will spend time making validating these variants and confirming their effects on gene function, others will simply assume that in a genome with 99.5% overall accuracy, it’s highly unlikely that such interesting variants will prove to be false. And in the high-pressure world of human disease genomics, they will be tempted to push their findings into the journals as quickly as possible before someone else beats them to that brand new disease gene.
So here’s the thing: the greater the predicted functional impact of a sequence variant, the more likely it is to be a false positive.
The reason for this will be pretty clear to the Bayesians in the audience (large-effect variants have a very low prior), but can take a while to fully appreciate for those without a natural statistical intuition. This effect occurs because variants with large effects on function are more likely to be harmful, so in general they are weeded out of the population by natural selection. In other words, the genome is highly depleted for variants with large functional effects.
Error, on the other hand, is more or less an equal opportunity annoyance – false positives, due either to DNA sequencing problems or issues with interpretation (e.g. thinking a region is protein-coding, when in fact it isn’t), appear without much regard for their effects on gene function.
So sequence changes with large effects on function are depleted for real variation, but have roughly the same overall error rate as the rest of the genome. As a result, the ratio of false changes to total observed changes – the false positive rate – is far, far higher for these variants.
This was a massive problem for our study, which looked at variants with about as large a predicted impact on function as it’s possible to get. As a result, we spent a very long time running experiments and designing filters, and ended up throwing away over half over the loss-of-function variants in our initial set: these discarded variants were either likely sequencing errors, apparent problems with gene annotation, or variants that simply didn’t look like they had much of an effect on gene function (for instance, gene-truncating variants that only remove the last 1 or 2 percent of a gene).
I should emphasise that a lot of these errors were the result of working with very early-stage sequencing data – some of the sequencing done for the pilot project is now over four years old. Modern sequencing data and variant-calling algorithms are far more accurate. In addition, as a result of this project and various other efforts gene annotation is now greatly improved – the Gencode annotation we use in the 1000 Genomes Project has been tweaked extensively over the last couple of years through the efforts of the HAVANA team and others. In fact, one of the few consolations of having to wade through a sea of errors was that we were able to fix many of them, so they won’t come up over and over again in future sequencing projects.
Still, while impressive, these are quantitative improvements, and the lesson stays the same: if you’re a PhD student working on large-scale sequencing data, and you find a fascinating mutation in your disease patient, be sure you validate the absolute hell out of that thing before you start drafting your paper to Science. The more fascinating it looks, the more you should disbelieve it – that’s as true in human genomics as it is in any other field of science.
What next?
So now we have a catalogue of LoF variants that we’ve stared at for a long time, and we’re now pretty sure that most of them are real. What can we actually do with this list? This is a question I’ll be pursuing pretty hard in my new position in Boston.
The first step is to make it bigger and better. The 1000 Genomes Project is rolling on – there are over 1,100 people now completely sequenced, and there will be some 2,500 by the end of the year – and we’re continuing our analysis of LoF variants as the project grows. But we’ve also set our sights on larger cohorts: for instance, the tens of thousands of exomes sequenced over the last couple of years in disease-focused projects. As we improve our filters and start applying them to human sequence data on this scale we expect to pretty rapidly build up a definitive catalogue of LoF variants present at any appreciable frequency in the population.
Then comes the exciting bit: figuring out what effect they might have on human variation and disease risk. The last paragraph of our paper hints at where this project might go:
Finally, we note that our catalogue of validated LoF variants comprises a list of naturally occurring “knock-out” alleles for over 1,000 human protein-coding genes, many of which currently have little or no functional annotation attached to them. Identification and systematic phenotyping of individuals homozygous for these variants could provide valuable insight into the function of many poorly characterized human genes.
Think of this as the Human Knockout Project – an effort take each gene in the genome and look for someone who has that gene completely knocked out. In some cases those people won’t exist – they will have died in utero. In other cases they will suffer from awful diseases – we already know of many such examples. But there will also be genes where eliminating function has more subtle or even beneficial effects. Surprisingly, we already know of several examples where rare LoF variants actually protect against disease, such as PCSK9 and heart disease, IFIH1 and type 1 diabetes, and CARD9 and Crohn’s disease – and these are very interesting to pharmaceutical companies seeking potential drug targets for these diseases.
We actually still know very little about the normal function of most genes in the human genome. If we can find individuals who are missing those genes, and take a close look at how they differ from the rest of the population, we’ll get clues that can then be followed up in downstream functional experiments.
This is a massively ambitious project, of course. We’ve made a start in this paper, by defining an initial list of LoF variants and coming up with filters that will help us track down more – but to really nail the effects of these variants will require a global, collaborative effort, (at least) hundreds of thousands of participants, and lots of dedicated researchers. Still, it can be done.
A personal note: the benefits of collaboration
I was very fortunate in this project to be able to work closely with some of the sharpest minds in human genomics – and without these collaborations this study would have been impossible. I owe a particularly huge debt of gratitude to Suganthi Balasubramanian from Yale and Adam Frankish from Sanger, who were my comrades in arms on this project from the beginning. The two senior authors on the paper, Mark Gerstein and my supervisor Chris Tyler-Smith, were also instrumental in shaping the project and driving it forward.
However, the author list for this paper is long for a good reason. When we wanted to look at the effects of LoF variants on common disease, I talked to Luke Jostins and Jeff Barrett, who know the area better than anyone else I know; to get a sense of their effects on gene expression, I talked to Joe Pickrell and Stephen Montgomery, who published back-to-back Nature papers on interpreting RNA sequencing data in 2010; to look at indels, I spoke to Zhengdong Zhang, Kees Albers and Gerton Lunter, while for structural variants I went to Klaudia Walter and Bob Handsaker. Others, like Ni Huang and James Morris, contributed heavily to multiple parts of the paper. Each of them brought enthusiasm and tremendous expertise to the project.
Research is never easy, but it’s certainly a lot more pleasant when you get to do it alongside people you like and respect.








 RSS
RSS Twitter
Twitter
Congratulations on your paper!
You obviously spent a lot of time manually rechecking and reannotating all the LOF variants to get at the real variants, which clearly required a ton of expert man hours.
Do you have any suggestions for how other groups could now leverage your work to improve annotations of their own data (without having to duplicate your heroic efforts!)? Would the most recent version of Gencode contain all/most of your revised annotations?
Hi Jessica,
Thanks!
Yes, you can get the benefit from our pain simply by using the latest Gencode release – if the HAVANA team spotted an error that could be fixed in this set, it has been. We’re also working on a more advanced annotation pipeline for LoF variants that will automatically filter out a lot of these problems, which should be ready later this year.
Great post and great paper! I couldn’t agree more with your conclusion that “rare LoF variants are where most of the action is in terms of effects on disease risk”. Regarding priors for severe mutations, do they increase if you are sequencing people with a severe disease? (I’m not arguing against the need for stringent validation, just suggesting that the ascertainment – from a person with disease or not – might change the prior).
great work. why not publish the findings in an open access journal!!! i think its time that we start providing equal opportunity to folks around the globe to have open and free access to the scientific output. after all, most of the science is funded by public money and why should public pay to have access to see the output of research!!! would you not agree? sorry, i don’t want to digress but this kind of great work should be published in open access journals. thank you.
Hi Kevin,
You’re right, the prior definitely goes up given that a serious disease patient is expected to contain at least one genuine disease-causing mutation. But as you say, this doesn’t justify skimping on validation!
Hi fairscientist,
As I mentioned in the post, you can download a pre-formatted version of the manuscript, along with all the supplementary data, here:
http://www.macarthurlab.org/lof
[This is permitted under Science‘s access policy.]
I completely agree that scientific findings and data should be made available as widely as possible. We currently live in a world where the journals that have the highest impact on the career progression of authors are closed-access. I think that’s wrong, and that it will shortly change, but right now it’s the reality we occupy – and it simply means it’s the responsibility of the authors who publish in these journals to ensure their data is made available via other means.
I look forward to the day that all scientific publications are freely available. Until that day, I’ll just have to do my best to ensure that the work we do is still accessible to the community: both by making the manuscripts and raw data freely available, and by blogging about it.
Well done on the paper Daniel and best of luck with the new job. I look forward to hearing how this work develops….
you said, “we currently live in a world where the journals that have the highest impact on the career progression of authors are closed-access” and that’s what we need to change. and i believe change is going to happen in opposing what we all believe in rather than maintaining the status quo. this way we bring the change that we desire rather than wish for something that, if left alone, will never happen in our lifetime. if you read michael eisen’s blog (http://www.michaeleisen.org/blog/?p=911), the academic jobs, grants and tenure are not solely awarded based on nature, science & cell publications and that refutes your pre-conceived notion that many young scientists hold.
anyway, thank you for posting the article.
@fairscientist, that’s like saying that money isn’t everything – it’s true, but try living with very little. Chances are it’s a much harder life. I think IF can be likened to academic currency – we may not like it, but it’s what almost everyone is using (whether it’s the cause of career progression or merely a very good proxy). I don’t think it’s reasonable to expect others to put themselves on the other side of the statistics just to make a point.
Daniel and colleagues have done some pretty amazing work with this project and they deserve to have it published in a high-impact paper like Science. The fact that he took the extra step to make it accessible to those without a subscription shows that he agrees with your belief.
I don’t know whether you or Michael Eisen are tenured scientists, but it’s far harder to live up to such beliefs when one isn’t (or when one doesn’t even head a lab yet, for that matter). This isn’t to say that you’re wrong, I agree that science (the endeavor, not the journal) should be openly accessible and the argument isn’t invalid merely because one has tenure or not. However I think that change should come from academic committees that accept new scientists and grant tenure, not from the young scientists who have put in 10+ years of hard work and risk losing it all just because they didn’t live up to the biases of those judging their work.
Wonderful study Daniel – you were namechecked and your study discussed at the MGH Scientific Advisory Committee talks today as an example of the sort of insight that is required for clinically relevant genome mining. Mark Daly clearly likes you….
Thanks mghOb – I was actually there in the meeting, and really appreciated Mark’s very kind words!
Hi Daniel, I had a chance to present this paper in our journal club and really liked it. The most interesting part of the paper seemed to come at the very end though, when you guys have tried to develop a linear discriminant model to see if all the genes can be scored. The model is not very good at the moment, but I pretty much liked the thought and intent behind the same.
I do have one comment though. To say that a normal healthy individual carries ~100 LoF genes is not quite true, I believe. As far as I understand, these results have emerged from the analysis of the sequencing data of NA12878 (the anonymous female of European ancestory). However, how accurately she represents the population is unclear. As a result, more studies devoted to the detailed analysis of individual genomes might be needed.
I was a bit surprised here because everywhere else in the paper, the caveats to any interpretation have been very well mentioned and discussed.