In a previous post I discussed copy number variation, a form of genetic variation not broadly reported by DTC companies. In today’s post I provide a very simple program that allows one to identify potential deletions on the basis of high density SNP genotypes from a parent-offspring trio, and report on the results of running this program on data from my own family.
The program uses an approach that I applied as a graduate student to mine deletions from the very first release of data from the International HapMap Project in 2004. The idea, explained in my last post, is to look for stretches of homozygous genotypes interspersed with mendelian errors, which might indicate the transmission of a large deletion. Let’s be clear, this is a simple analysis that most programmers and computational biologists would find straightforward to implement. It is probably a good practice problem for graduate students and would-be DIY personal genomicists.
I obtained 23andMe data from both my mom and dad, and, with their consent, ran the three of us through the program. I was mildly surprised to find only two potential deletions; I had previously speculated that one would find 5-10 deletions per trio with the 550K platform used by 23andMe.
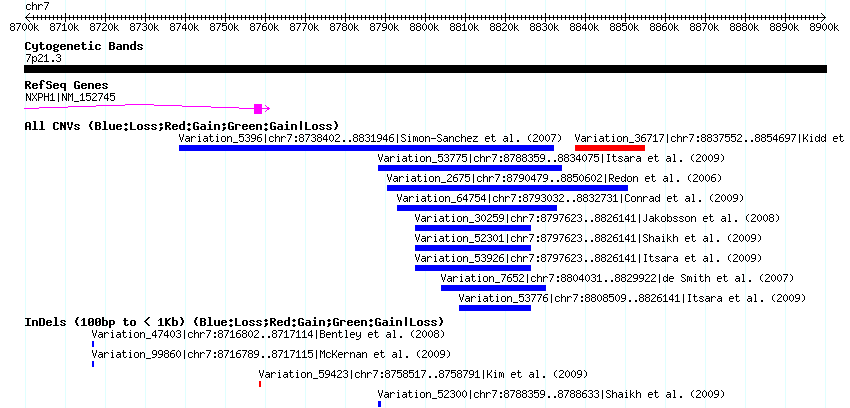
These two deletions were dramatically different in size and level of support. The first, in a non-coding region on chromosome 7, appears to be about 50kb in size (the first SNP outside of the deletion on each end is rs10228390 at 8788633 and rs917038 at 8839252), contains 18 SNPs compatible with a deletion transmission, and 9 of these SNPs (a full 50%) are “Mendelian errors” (meaning that they are not consistent with the pattern of transmission you would expect to see from parent to child). Furthermore, one can confirm that this deletion is common and has been previously identified by searching the Database of Genomic Variants (click image to zoom):
The second deletion is on chromosome 15, between 31505941 bp and 31519908 bp, falling within an intron of the gene RYR3 , a brain ryanodine receptor. This putative deletion is no more than 14kb in size and spans only 3 SNPs, 2 of which are mendelian errors. Again looking at DGV, there are CNVs that have been reported to span this region, however they are much, much larger (>150kb), reflecting the lower resolution approaches used in those studies. No variant has been reported at this locus with a high-resolution technology (like next-sequencing or high-res oligo arrays) so it is difficult to say if this is a known variant or not.
There are two principle explanations for the low yield of deletions from these data. One is statistical sampling: the number of large deletions in my family is just on the low end of the spectrum of all the families out there. The second, and more appealing to my intuition, is that in the quest for extremely high quality SNP genotypes, 23andMe has (appropriately) replaced many of the SNP genotypes in regions of deletions with missing data.
In order to test this latter hypothesis, I asked the question, is there evidence for an enrichment of missing data in regions of known, common deletions, in my own data? I was impressed to see that, across the entire set of SNPs, I had only 1453 missing data points, a call rate of 99.75%. I made a list of 1,008 deletions that were recently reported to have a population frequency greater than 5% in Europeans. Out of 975 SNPs that fell within the boundaries of these common deletions, 84 (8.6%) were “no calls”, a massive enrichment over the proportion for the total dataset (0.25%).
For those of you with data from trios (two parents and a child) who would like to look for deletions in your own genome, I’ve posted some code and directions here. In coming weeks I hope to make the code easier to run, and also add a test of the presence of known, common deletions by looking for excess homozygosity or missing data. Given some reference files that contain the location and frequency of known deletions, as well as allele frequencies for all SNPs in these regions, it should be possible to pull out large stretches of homozygosity that are statistically improbable from a copy-neutral (non deleted) genome. This is something that can be applied to single genomes (no families required!). Finally, there are other, sophisticated approaches for finding deletions out there now, that work on SNP genotype data from unrelated individuals, which others may want to explore:
Gusev, et al. (2009) Whole population, genome-wide mapping of hidden relatedness








 RSS
RSS Twitter
Twitter
Paid promethease runs have a feature where parent child genomes can be compared to do *some* of the sort of analysis described above. It doesn’t name the regions, but I suspect the deletions described above should be readily visible in the images. This is better explained at
http://www.snpedia.com/index.php/Promethease/Features#Genomic_similarity
I’m be grateful to know if his missing regions are readily apparent in
the graphs.
Very interesting – but I’m slightly unclear as to whether you think the “no calls” in common deletions are personal to you (i.e. you might have inherited a deletion from both parents, and so have nothing to report at that site); or are systematically applied by 23andme, because they know these SNPs give odd results (and we now know this is because they fall in common deletions).
With regards whether the failure-to-inherit a small chunk of DNA would necessarily matter to you, healthwise, a recent paper:
Pelak, K et al (2010)
The characterization of twenty sequenced human genomes
PLoS Genetics 6(9) e1001111
http://www.plosgenetics.org/article/info%3Adoi%2F10.1371%2Fjournal.pgen.1001111
suggests that complete loss of function of a gene is commonplace – they suggest we have, on average, 165 of these each. Remarkably resilient organism, the evolved mammal.
@Neil
Are mammals more tolerant of large coding deletions than other animals/organisms?
Hi Neil,
With 12 separate 23andMe files within the group (and several dozen more available from various sources) we should be able to untangle the “generic filtering” vs “no-calls due to personal CNVs” hypothesis.
Regarding the numbers from the Pelak et al. paper: I’ll be presenting the results of large-scale experimental validation and reannotation of gene-inactivating variants from 1000 Genomes at ASHG in a couple of weeks. Long story short, while humans do have more genuine gene losses than we might have intuitively expected, the numbers reported so far (including from the Pelak paper) are heavily inflated by sequencing and annotation errors. This is exactly what you would predict given the relatively low prior on a gene-inactivating variant being real – Chris Tyler-Smith and I discuss this in a recent HMG review.
I noticed some oddities in my ROH, R_ROH workup for the Amish/Mennonite/Brethren project based upon David Pike’s code. I have 2 long parent-child ‘deletions’ in my multi-generational family set. I have not gotten around to looking at the small single SNP changes to see if they represent CNV variants. I look forward to your abusing your code.
Note that I run 64-bit OS’s so hopefully your code will function outside of the 32-bit universe.
@Wayne – I’ve just posted the code to https://genomesunzipped.org/data. Good luck, will be interested in how you get on.
@Neil – I don’t believe that 23andMe are a priori masking genotypes from regions of known common CNVs. The missing data phenomenon is more likely a by-product of the statistical methods used to generate genotypes. Typically SNP genotyping algorithms try to cluster the raw “intensity” data generated from the SNP chip into discrete groups corresponding to one of three genotype predicted to exist at the locus (AA, AB, or BB). Obviously when you have a CNV segregating at a SNP locus, then you have more than three possible genotypes (ie A-/B-/–, plus the original three). So samples with CNVs will appear as outliers to copy normal clusters (that is they will have lower probability of being a member of any given cluster). I believe 23andMe uses a stringent cutoff for this assignment probability, to reduce genotyping error at copy normal sites, and as a result deletion carriers are more likely to be set to “NN”, even if they are heterozygous for the deletion.
@Don – I have Cygwin ready under Windows by no Delscan.c file was present in the archive….
@Wayne – sorry about that… it should be there now!
@Don You should specify which Linux distro(s) and GCC version your binaries are compiled for. They seem to be somewhat unhappy under the latest Ubuntu and g++ versions. I have a VMware image for that environment.
e-mail me…. wkauffman (-at-) yahoo (.dot.) com . Need to work a few things out. Need to add support for multiple children.
Under Cygwin Windows getting 6 output lines for Chr 8 test data and you indicate it should be 13.
Recorded 3 pedigree entries
Read 1 Families
Read 3 samples from snp file
Read 77111 loci from snp file
First seqname BOB Last Findiv RUPERT
First Locus 151222 Last Locus 146264218
Hi Wayne,
From what you’ve shown me, it looks like you have everything working! The text that you have pasted into the post are status updates printed to the screen by the program, not the results per se. As described in the README, running the perl script on the example data “should produce a file called ‘hapmap.fam.dels’ containing 13 deletion intervals”. You should be ready to look at your own data now, by creating a new family file for each trio that you want to analyze.
I’ll send you an email and we can discuss offline.
Cheers
Don
Hi,
Here are a few questions for the group:
a) Does a stretch of — calls in the raw 23andMe data files indicate a high probability of a deleted region?
b) How common is it to have a long stretch of homozygous calls (perhaps more than 100 in a row)? Is this a fairly common occurrence?
Thanks,
Michael