To celebrate the end of the blogging year here at Genomes Unzipped, we wanted to spend a bit of time reminiscing about the papers we enjoyed the most in 2010. Feel free to add your own suggestions in the comments!
Joe: Mice, men, and PRDM9. A key goal in evolutionary biology is to identify the mechanisms leading to speciation. One way to get at that goal is to identify genes that cause sterility or reduced fitness in hybrids between species or diverged populations. In mammals, exactly one such gene has been identified to date: the DNA-binding protein PRDM9. This year, three groups working on a seemingly different problem–deciphering the molecular mechanisms by which recombination shuffles genetic variation between generations–stumbled across an important gene in this process: PRDM9. Variation in this gene influences recombination patterns in both mice and humans, and is responsible for the dramatic differences in recombination patterns between humans and chimpanzees. Is it a simple coincidence that a gene which influences recombination also appears to have a role in speciation? Time will tell.
Parvanov et al. (2010) Prdm9 Controls Activation of Mammalian Recombination Hotspots. Science. DOI: 10.1126/science.1181495.
Baudat et al. (2010). PRDM9 Is a Major Determinant of Meiotic Recombination Hotspots in Humans and Mice. Science. DOI: 10.1126/science.1183439.
Myers et al. (2010). Drive Against Hotspot Motifs in Primates Implicates the PRDM9 Gene in Meiotic Recombination. Science. DOI: 10.1126/science.1182363.
Daniel: Whole-genome sequencing to develop personalised cancer assays. The area of medicine where the transforming power of new DNA sequencing technologies is moving the fastest is in cancer diagnostics and therapy. There were many studies relevant to this field in 2010 (with a fair proportion featuring on the excellent MassGenomics blog), but this paper was a simple, elegant example: the authors performed low-coverage whole-genome sequencing of four tumour samples, identified large genomic rearrangements present in the tumour cells but not in the patient’s healthy tissue, and then designed personalised, quantitative assays measuring the proportion of cells carrying these rearrangements in the patients’ blood. These assays allowed them to track, almost in real time, how the patients’ cancers responded to various therapies, like so:
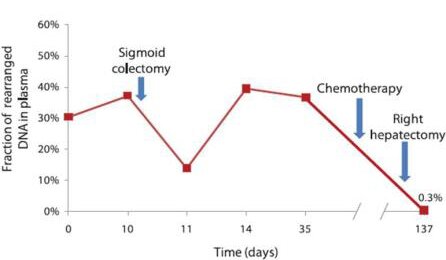 Leary et al. (2010) Development of personalized tumor biomarkers using massively parallel sequencing. Science Translational Medicine. DOI: 10.1126/scitranslmed.3000702.
Leary et al. (2010) Development of personalized tumor biomarkers using massively parallel sequencing. Science Translational Medicine. DOI: 10.1126/scitranslmed.3000702.
Joe: Wasabi and rattlesnake heat vision. One way in which high-throughput sequencing technologies are changing the way biology is done is by opening up the possibility of genome-scale analyses in non-model organisms. This year, nothing exemplified this better than a paper identifying the protein TRPA1 (which, in humans, triggers the “hot” sensation you get from eating wasabi) as the molecule used by rattlesnakes and pythons for infrared vision. The first step for these authors was an RNA sequencing study of the genes expressed in the rattlesnake pit organ (known to be the organ used to sense heat). A few years ago, this sort of thing would have been unthinkable.
This paper also gets bonus points for the understatement of the year: “Snakes, particularly pit vipers, are inconvenient subjects for physiological and behavioural studies”. Indeed. Always have this paper on hand when your mouse geneticist friends complain about getting scratched or bitten by their subjects!
Gracheva et al. (2010) Molecular basis of infrared detection by snakes. Nature. doi:10.1038/nature08943.
Vincent: finding the viral triggers for autoimmune disease. For many diseases the trigger is likely to be a combination of genetic and environmental factors. However, this knowledge is typically hard to obtain: while genetic factors can be measured in large populations, we usually lack the knowledge of the environmental factors. In this paper, the authors take a direct route using an animal model to show that a combination of a viral infection on a specific genetic background triggers a reaction that looks very much like Crohn disease. What strikes me with this paper is that the finding is important but the discovery was complex and this result could have been missed for many years without this remarkable work. The finding should be confirmed of course by other groups but if it turns out to be correct, it suggests plenty of new studies to better understand autoimmunity and get us closer to treatments. Future work also includes genetic analyses to move away from the mouse model and understand the role of these associated variants in human populations.
Cadwell et al. (2010) Virus-plus-susceptibility gene interaction determines Crohn’s disease gene Atg16L1 phenotypes in intestine. Cell. DOI: 10.1016/j.cell.2010.05.009
Jeff: a retort to the GWAS-haters. 2010 was marked by both bigger and bigger GWAS studies which uncovered hundreds of new associations to common diseases and by a rising trend to dismiss such studies as pointless wastes of time and money. On the one hand researchers are finding regions of the genome associated with an ever-increasing list of traits and diseases, on the other hand these effects are generally individually tiny and without much use in predicting disease risk. Of course, those of us who have been working on GWAS know that disease prediction is only one side of the GWAS coin and that the other side (namely, understanding biology) is almost certainly more important.
The lovely SORT1 paper (a companion to a ginormous GWAS which brought the tally of lipid loci to 95) is one of the first of what I expect to become frequent post-GWAS genomic biology papers over the next few years. The authors show how a GWAS hit SNP creates a transcription factor binding site and thus alters expression of SORT1 in the liver. They go on to show with siRNA knockdown and viral overexpression experiments in mouse that changes in the gene’s expression affect lipid levels. A really excellent example of how GWAS results have opened up a whole new area of biological and medical research.
Musunuru et al. From noncoding variant to phenotype via SORT1 at the 1p13 cholesterol locus. Nature. 2010 Aug 5;466(7307):714-9.
Luke: connecting heritability, familial risk and genetic risk prediction. My favourite paper this year, from Naomi Wray et al, did not contain any new groundbreaking discovery, but instead tied together some key results that underlie twin studies and the calculation of heritability, measurements of familial risk, and the sort of measurements that we use to assess genetic prediction of disease. The outcome of this were some simple methods (and a simple online calculator) to convert between heritability, sibling recurrence risk (how much more likely you are to develop a disease if your sibling does), and the AUC (a measure of prediction). These are all particularly relevant to personal genomics, and the calculations in this paper underline the fact that if we could explain all the heritability we see in twins, we could also make clinically useful predictions of disease in unrelated individuals.
Wray et al (2010) The Genetic Interpretation of Area under the ROC Curve in Genomic Profiling PLoS Genetics DOI:10.1371/journal.pgen.1000864








 RSS
RSS Twitter
Twitter
My favorite paper was the publication of the Neandertal genome which shed new light on the history of non-African humans.
Hi Luke,
With regard to the heritability paper, the point of it is that if the prevalence of the disease is low and the heritability in liability scale is high, then we only need to explain a fraction of the heritability to make a 23andme-like risk prediction test useful in a clinical setting. For example, in the paper, only 10% of the 0.64 heritability needs to be explained for Lupus to achieve a clinically useful 0.75 AUC.
@GftE
I wasn’t really convinced by the argument that an AUC of 0.75 defines “clinical usefulness”. For something as rare as Lupus, an AUC of 0.75 would not be very useful for screening, which is the first thing most poeple will think of when they think of clinical usefulness. However, factors with pretty low effect size can still be clinically – the AUC of smoking is in the region of 0.6, but doctors can certianly still use that information when diagnosing lung cancer.
However, the sort of AUCs given if you can capture all heritability (most greater than 0.9) are unambigously very useful in many circumstances. For instance, a Crohn’s screening test, using the full heritability, would be able to identify 75% of people who have Crohn’s, and only follow up 2-3 false positives for each true positive, which would be very impressive for a rare disease.